Show code cell content
import matplotlib.pyplot as plt
import numpy as np
import SimpleITK as sitk
from armscan_env import config
from armscan_env.clustering import TissueClusters
from armscan_env.envs.rewards import anatomy_based_rwd
from armscan_env.envs.state_action import ManipulatorAction
from armscan_env.util.visualizations import show_clusters
from armscan_env.volumes.loading import (
load_sitk_volumes,
normalize_sitk_volumes_to_highest_spacing,
)
from armscan_env.volumes.volumes import TransformedVolume
config = config.get_config()
Volume Random Transformations#
Let’s load all the volumes in their original shape and in the normalized shape:
volumes = load_sitk_volumes(normalize=False)
normalized_volumes = normalize_sitk_volumes_to_highest_spacing(volumes)
print(volumes[0].GetSize())
print(normalized_volumes[0].GetSize())
(606, 864, 61)
(771, 1100, 61)
Now you can set as volume any of the volumes(normalized or not):
volume = normalized_volumes[2]
volume_img = sitk.GetArrayFromImage(volume)
size = np.array(volume.GetSize()) * np.array(volume.GetSpacing())
print(f"{size=} mm")
transversal_extent = (0, size[0], 0, size[2])
longitudinal_extent = (0, size[1], 0, size[2])
frontal_extent = (0, size[0], size[1], 0)
size=array([159.83333606, 223.02778158, 69.99993324]) mm
optimal_action = volume.optimal_action
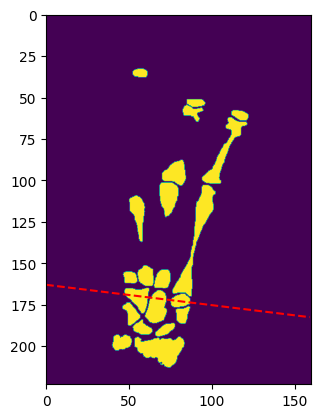
plt.imshow(volume_img[41, :, :], extent=frontal_extent)
o = volume.GetOrigin()
x_dash = np.arange(size[0])
b = volume.optimal_action.translation[1]
y_dash = x_dash * np.tan(np.deg2rad(volume.optimal_action.rotation[0])) + b
plt.plot(x_dash, y_dash, linestyle="--", color="red")
plt.show()
sliced_volume = volume.get_volume_slice(
action=volume.optimal_action,
slice_shape=(volume.GetSize()[0], volume.GetSize()[2]),
)
sliced_img = sitk.GetArrayFromImage(sliced_volume)
print(f"Slice value range: {np.min(sliced_img)} - {np.max(sliced_img)}")
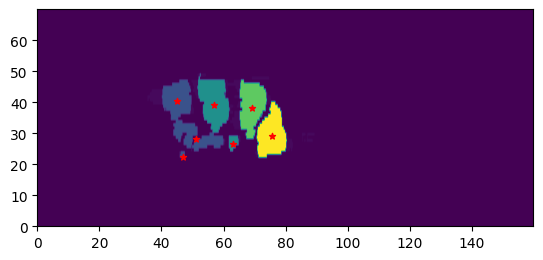
cluster = TissueClusters.from_labelmap_slice(sliced_img.T)
show_clusters(cluster, sliced_img.T, extent=transversal_extent)
reward = anatomy_based_rwd(cluster)
print(f"Reward: {reward}")
plt.axis("off")
plt.show()
Slice value range: 0 - 3
Reward: -0.041666666666666664

volume_transformation = ManipulatorAction(
rotation=(-7.213170270886784, 0.0),
translation=(-7.31243280019082, 9.172539411055304),
)
transformed_volume = TransformedVolume(volume, volume_transformation)
transformed_action = transformed_volume.optimal_action
print(f"{volume.optimal_action=}\n{transformed_volume.optimal_action=}\n")
volume.optimal_action=ManipulatorAction(rotation=(7, 0.0), translation=(0, 163))
transformed_volume.optimal_action=ManipulatorAction(rotation=array([14.21317027, 0. ]), translation=(0, 156.58286047804256))
transformed_img = sitk.GetArrayFromImage(transformed_volume)
plt.imshow(transformed_img[40, :, :], extent=frontal_extent)
ot = transformed_volume.GetOrigin()
x_dash = np.arange(size[0])
b = transformed_action.translation[1]
y_dash = x_dash * np.tan(np.deg2rad(transformed_action.rotation[0])) + b
plt.plot(x_dash, y_dash, linestyle="--", color="red")
plt.show()
sliced_transformed_volume = transformed_volume.get_volume_slice(
action=transformed_action,
slice_shape=(volume.GetSize()[0], volume.GetSize()[2]),
)
sliced_transformed_img = sitk.GetArrayFromImage(sliced_transformed_volume)
print(f"Slice value range: {np.min(sliced_transformed_img)} - {np.max(sliced_transformed_img)}")
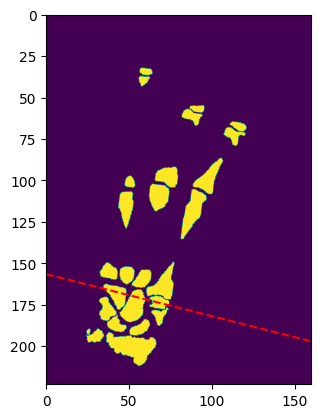
cluster = TissueClusters.from_labelmap_slice(sliced_transformed_img.T)
show_clusters(cluster, sliced_transformed_img.T, extent=transversal_extent)
reward = anatomy_based_rwd(cluster)
print(f"Reward: {reward}")
plt.show()
Slice value range: 0 - 3
Reward: -0.041666666666666664